Recently, while scoping the brave new world of precision oncology, we were introduced to somatic mutations as a key culprit in cancer formation. Let’s delve a little deeper to see why they are so important.
Somatic variants are mutations that occur in any body cell except germ cells, meaning they are not heritable. These mutations are acquired through natural cellular processes like cell replication and aging, or from biochemical stressors such as smoking, infection, and irradiation. Although complex DNA error repair mechanisms usually prevent these changes, some variations may slip through undetected [1].
Most cancers originate from such somatic mutations, which is why they are often localized to specific organs or tissues, and are called sporadic cancers [2].There are two types of somatic variants: ‘driver’ mutations, which promote accelerated cell proliferation and are the main causal force behind the cancer, and ‘passenger’ mutations, which are present in cells that clonally expand to form the tumor but do not drive tumor growth [1]. Due to their prevalence and localized nature, and the promising results of targeted treatments, somatic mutations have become a significant focus of research.
Large scale sequencing projects from the early 2000s, such as The Cancer Genome Atlas (TCGA), Cancer Genome Project (CGP), and Pan-Cancer Analysis of Whole Genomes project (PCAWG), have characterized over 20,000 primary cancers and matched normal samples across more than 38 cancer types. Thousands of somatic variations are now recorded, even within a single cancer genome [3].
Most somatic mutations are ‘passengers’, but research has unpacked a surprising number of them that do drive cancer. Further, many sequencing parameters such as read depth and alignment software affect accurate variant calling. Identifying cancer ‘driver’ mutations and understanding their impact on the tumor is now a central requirement in cancer genomics [4].
Precision oncology categorizes somatic mutations based on clinical actionability, i.e., the availability of effective therapy. However, clinical evidence for genetic variants from literature is often scattered, and the clinical significance of many novel or rare variants remains uncertain. The rapid development of new targeted drugs also challenges clinicians to stay updated, sometimes preventing critical information from reaching patients when needed [5].
Enter databases. They address this issue by utilizing massive computing power to store and organize thousands of somatic mutations, making them accessible instantly through search functions. Each variant is annotated by experts with details on its impact on genes, proteins, and associated pathways. User-friendly tools help clinicians and researchers find clinical implications and targeted drug information. Regular updates ensure accuracy and relevance.
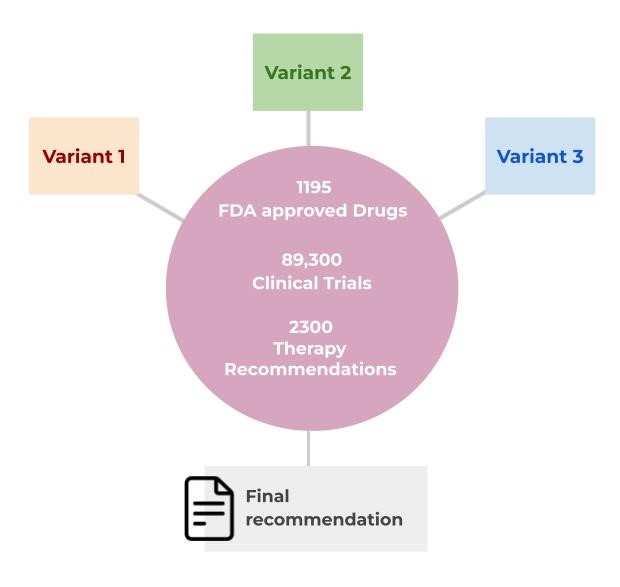
While several large databases like CGI, CIViC, COSMIC, JAX-CKB, and OncoKB offer such platforms, they are not without flaws. Some may lack comprehensive therapy information, while others can be costly, infrequently updated, or difficult to navigate. Ideally, a richly curated and frequently updated somatic variant database would integrate seamlessly with a variant classification system and provide comprehensive treatment-related information for any detected variant. The Strand Somatic Cancer Knowledgebase is designed to offer just that, maintaining over 640 genes, 24,000 variants, 1195 cancer-related targeted drugs, 89,300 clinical trials, and 2300 therapy recommendations.
Together with the StrandIris platform, it can systematically figure out whether a variant, or a combination of them, is culpable for cancer, and how best to go about treating it. We shall explore how in the next blog of the series.
You can find more details regarding StrandIris, here!
Explore the rest of the blog series below:
- Part 1: Precision Oncology: A Genetics Revolution in Cancer Management
- Part 3: Somatic Variant Classification—Mining for Clues
- Part 4: Somatic Variants: Towards Better Therapy
References:
- Greenman, C., Stephens, P., Smith, R., Dalgliesh, G. L., Hunter, C., Bignell, G., & Stratton, M. R. (2007). Patterns of somatic mutation in human cancer genomes. Nature, 446(7132), 153-158.
- Germline and Somatic Variants: What Is the Difference?
- Sondka, Z., Dhir, N. B., Carvalho-Silva, D., Jupe, S., Madhumita, McLaren, K., ... & Teague, J. (2024). COSMIC: a curated database of somatic variants and clinical data for cancer. Nucleic Acids Research, 52(D1), D1210-D1217.
- Chen, Z., Yuan, Y., Chen, X., Chen, J., Lin, S., Li, X., & Du, H. (2020). Systematic comparison of somatic variant calling performance among different sequencing depth and mutation frequency. Scientific reports, 10(1), 3501.
- Gao, P., Zhang, R., & Li, J. (2019). Comprehensive elaboration of database resources utilized in next-generation sequencing-based tumor somatic mutation detection. Biochimica et Biophysica Acta (BBA)-Reviews on Cancer, 1872(1), 122-137.