Early cancer detection and personalised treatments are crucial advances in precision oncology. Liquid biopsies (LBs) play a significant role in this, offering a minimally invasive way to detect cancer and tailor therapies for patients. Another important advantage of LBs is that they can capture tumor heterogeneity and provide insights into tumor genomics, making them invaluable for stratification and monitoring of cancers.
Biologically, liquid biopsies analyse both, cellular structures such as circulating tumor cells, extracellular vesicles, and tumor-educated platelets and cell-free molecules including cfDNA, cfRNA, proteins, metabolites, and lipids. However, developing a liquid biopsy assay is a complex process. Early-stage cancers release very low levels of tumor DNA and require highly sensitive and specific bioinformatics pipelines to detect low-frequency variants.
Recently, Strand supported a liquid biopsy diagnostic company developing a non-small-cell lung cancer (NSCLC) assay with an aggressive time-to-market goal, achieving complete implementation in just 3 months. The project resulted in a successful product launch and a 70% reduction in costs, lowering sequencing expenses from $20/GB to $6/GB, primarily due to savings in sequencing and cloud infrastructure. The client required scalable bioinformatics workflows, tracking of variant allele frequencies across multiple mutation types, integration of analysis tools, and validation processes to support regulatory compliance.
To meet these requirements, we built an integrated bioinformatics pipeline for high-sensitivity NGS assays capable of detecting variants with allele frequencies as low as 2-5%.
An automated workflow processed sequencing data through primary analysis, secondary analysis for alignment and variant detection, and clinical report generation on a secure cloud-based platform. We ensured assay reliability through experimental validation and computational methods. Overall, the workflow streamlined data transfer, improved consistency, reduced processing time, and was documented to support CLIA compliance while being scalable for future patient analytics.
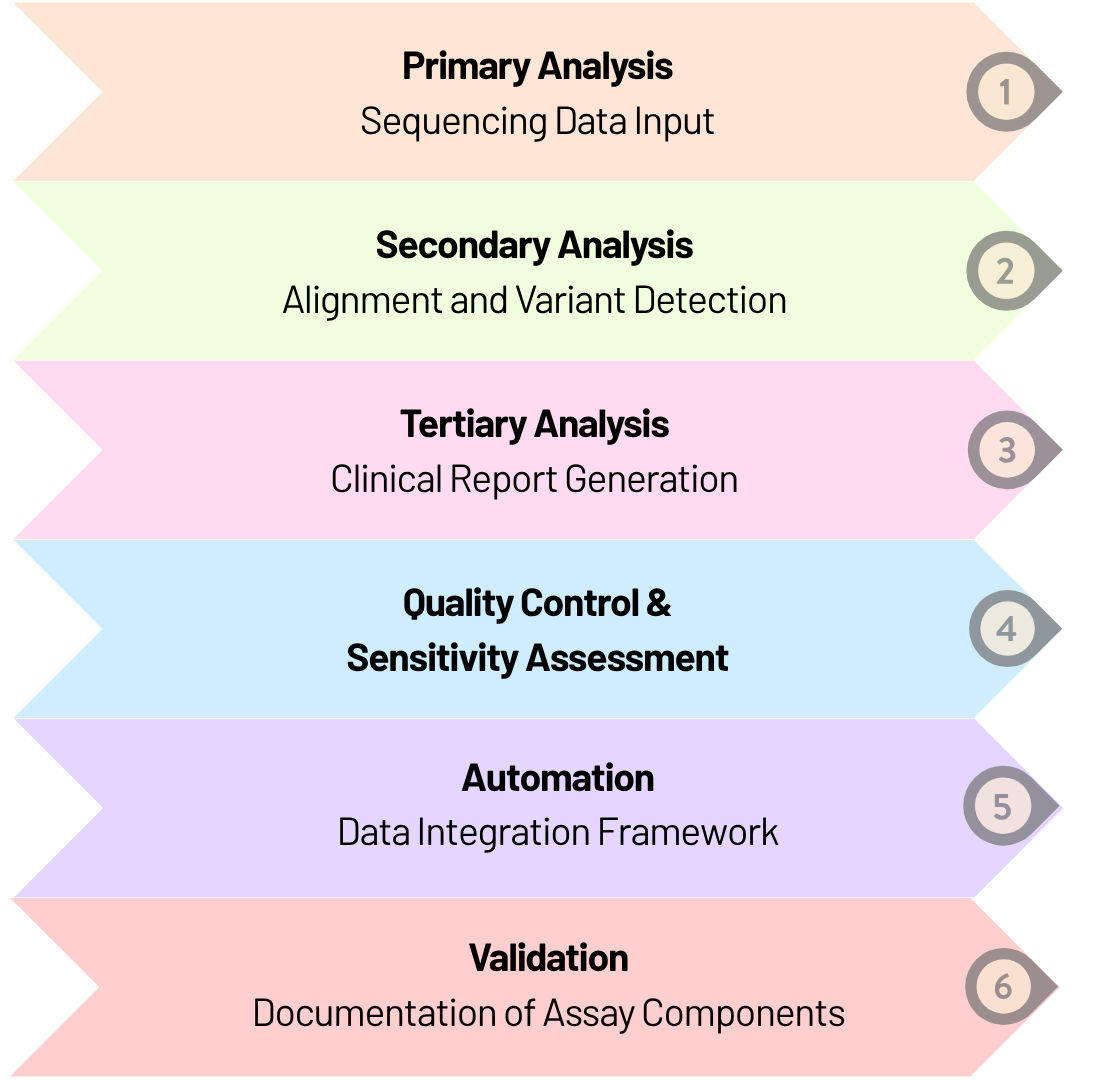
In summary, this work reduced assay development turnaround time for the client and enabled a timely market launch. It also demonstrates the critical role of bioinformatics in supporting scalable and efficient liquid biopsy assays, with potential applications across other cancer types.
To learn more about developing bioinformatics solutions for liquid biopsy assays, read our case study or write to us at [email protected].
References:
- Di Sario G, Rossella V, Famulari ES, Maurizio A, Lazarevic D, Giannese F, Felici C. Enhancing clinical potential of liquid biopsy through a multi-omic approach: A systematic review. Front Genet. 2023 Apr 3;14:1152470. doi: 10.3389/fgene.2023.1152470. PMID: 37077538; PMCID: PMC10109350.
- Tivey A, Lee RJ, Clipson A, Hill SM, Lorigan P, Rothwell DG, Dive C, Mouliere F. Mining nucleic acid "omics" to boost liquid biopsy in cancer. Cell Rep Med. 2024 Sep 17;5(9):101736. doi: 10.1016/j.xcrm.2024.101736. PMID: 39293399; PMCID: PMC11525024.