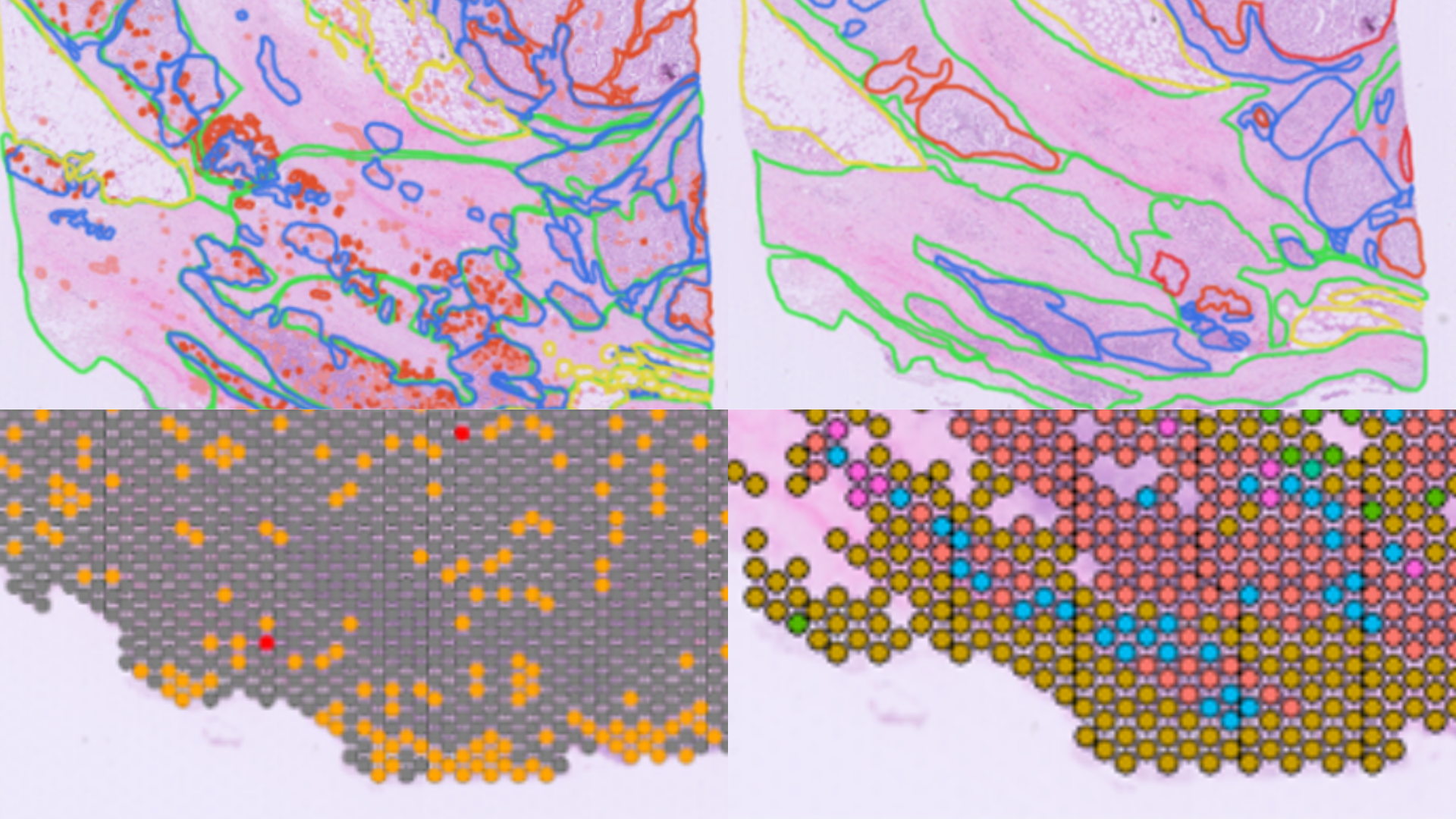
Recently, we explored the exciting field of targeted protein degradation during our webinar, “Unlocking New E3 Ligases for Targeted Protein Degradation in Cancer Therapy.” Here’s a quick overview.
The talk opened up with an exploration of PROTACs (proteolysis targeting chimeras) and molecular glues—two essential tools that employ the ubiquitin-proteasome system (UPS) for the degradation of proteins of interest. This system is key for tagging and breaking down unwanted proteins. By targeting specific proteins that promote cancer, these technologies offer a focused way to stop cancer effectively and selectively, opening up new possibilities for treatment.
A key focus of our discussion was on E3 ligases, the proteins responsible for selecting other proteins to be broken down. Although there are over 600 types of E3 ligases, only a few, such as CRBN and VHL, are commonly used in treatments. This limited use can result in resistance, a significant challenge in cancer therapy. The discussion emphasized the critical need to expand our use of E3 ligases to more effectively address this problem.
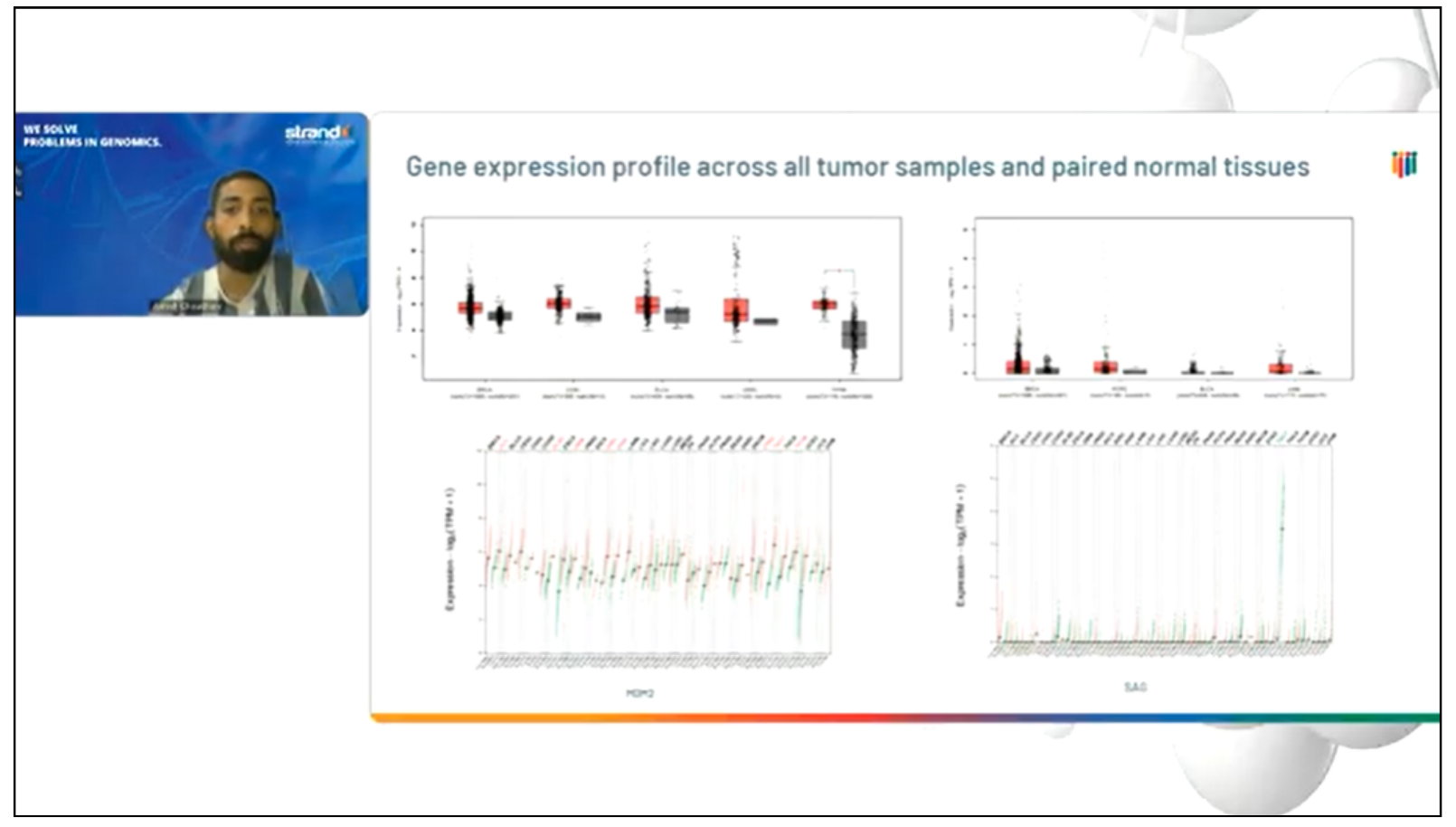
Our presenter, Ashish Kumar Choudhary, a scientist at Strand and an expert in genomics data analysis and bioinformatics, discussing the results of the study.
Among the most exciting parts of our discussion was the introduction of new and specific E3 ligases like c10orf90. This ligase is notable for its selective binding, especially with key proteins like TP53, a vital cancer regulator. This selectivity could lead to treatments that are more effective and have fewer side effects. We contrasted c10orf90 with broader-spectrum E3 ligases, which may cause unintended effects because of their wide range of interactions.
At Strand Life Sciences, we are advancing the development of novel cancer therapies by using our sophisticated bioinformatics tools and custom curation services. Databases like UbiHub and UbiBrowser2.0 help us identify E3 ligases that are overexpressed in specific cancers—crucial information for developing targeted protein degradation (TPD) strategies.
For deeper insights and support in your drug development efforts, read our latest case study and solution snapshot.