Plant genomics and transcriptomics are steadily reshaping the way we study crops. With genomics providing the genome sequence and transcriptomics showing which genes switch on or off as conditions change, we can now gain a much clearer view of how plants respond to drought, pests, soil variation, and other pressures that influence yield and resilience. This perspective supports efforts to develop crops that can withstand a changing climate and other challenges.
Plant genomes are often difficult to study because many are very large, contain long stretches of repetitive DNA (commonly due to transposons), or carry several sets of chromosomes (polyploidy). This makes assembly and analysis challenging and drives the need for higher resolution sequencing tools and stronger data analysis methods.
As sequencing tools have improved, plant research has expanded far beyond single gene studies. Researchers can now work with complete genomes, expression profiles, epigenetic patterns, and data from entire populations.
What is driving the shift from microarray to NGS?
Since the early days of plant genomics, microarrays have been the standard tool for measuring gene expression. Their affordability and practicality made them widely adopted, and they remain useful in some settings. As sequencing technologies have advanced, however, certain limitations have become clear. Microarrays rely on known probes, which restricts their ability to detect new transcripts or unexpected genetic variation. They also have a limited dynamic range, so subtle differences in expression may not be captured.
High throughput sequencing is gradually becoming the preferred approach as it overcomes many of the constraints seen with microarrays. By reading sequences directly, it can reveal new genes, isoforms, structural variants, and single nucleotide changes that would otherwise go undetected. As sequencing costs fall and workflows become more manageable, more laboratories are able to bring sequencing into their routine studies.
Why Cloud-based Analysis is Becoming Essential
Modern sequencing produces datasets that are often too large for local machines to handle. Large genome assemblies, long read datasets, and pan-genome projects require storage and compute capacity that many labs cannot support on site. As a result, cloud platforms have become a practical way to run heavy workflows, share data across research groups, and scale analyses.
Supporting Technologies: Long Reads and Pan-Genomes
Many horticultural species have complex genomes shaped by hybridization and domestication. Often, this complexity cannot be resolved by short reads alone, but in combination with long reads, we get stronger, chromosome level assemblies that improve annotation, trait mapping, and variant discovery.
A single reference genome cannot capture the full diversity within a species. Some genes are shared across all varieties, while others appear only in certain lines and influence traits such as stress tolerance, yield, or local adaptation. Pan-genomes capture this broader range of variation and help identify traits that would be missed with a single reference. These studies depend on large sample sets and substantial compute, which suits cloud based analysis.
This approach also benefits orphan and underused crops. Many regions rely on local species with limited genomic resources, and new sequencing methods now allow them to be studied with the same depth as major crops.
What These Advances Enable
- Better trait discovery: Improved assemblies and more accurate variant calls lead to stronger genome-wide association studies, and better inputs for genomic selection. These insights help guide breeding for resilience and productivity.
- Integration of different data layers: Modern studies often combine DNA sequences, RNA expression, epigenetic patterns, and metabolite profiles. These integrated datasets give a more complete view of how traits arise. They also support precise gene editing with tools such as CRISPR, base editing, and prime editing.
How Strand Supports Plant Genomics Research
Our team has carried out genomics and transcriptomics studies in more than sixteen plant species over the past three years. These include fruits, vegetables, crops, millets, and even insectivorous plants. We work with newly assembled genomes, multi-omics studies, and complex datasets. This includes one of the largest plant genomes, onion, with an estimated size of 16 Gb.
We use both open source tools and our dedicated platform, Strand NGS. The platform supports DNA, RNA, and microbial sequencing data and helps run complete workflows from raw data to analysis and visualization.
|
RNA-Seq Analysis Phase |
Strand NGS Capacities |
Application in Plant Genomics |
|
Pre-Processing & Alignment |
Read/Sample filters, Align reads, Discard poorly aligned reads, Check Gene Body Coverage |
Handles alignment for large genomes; crucial for managing samples with high novel events. |
|
Variant and splicing analysis |
SNP analysis - detection and advanced filters to get high confidence SNPs, Splicing analysis and Gene Fusion detection |
Help uncover genetic and regulatory changes that drive key plant traits, including stress tolerance, development, and overall crop performance. |
|
Quantification & Normalization |
Quantify Gene Expression, Normalize Gene Expression, Filter genes expressed at low levels |
Ensures accurate quantification for large DE comparison sets (e.g., see DEG comparison in the foxtail millet study webinar) |
|
Advanced Statistics |
Statistical tests, Confounding Variable Analysis, Correlation, Class Prediction |
Facilitates complex cross-tissue (Leaf vs. Root) and cross-genotype comparisons. |
|
Downstream Analysis, Visualization & Reporting |
PCA, t-SNE, UMAP, Cluster entities/samples, volcano plots, violin plots, Gene Ontology, network and pathway analysis |
Generates visually compelling and biologically informative outputs (e.g., see PCA and correlation plots in the foxtail millet study webinar). |
Working With Difficult Genomes
A key strength of Strand NGS is its ability to handle complex alignment cases that often cause open-source tools to fail. For samples with a high frequency of novel events, such as onion, the platform uses a custom, scalable solution that splits, aligns, and merges reads to overcome memory and thread limitations and maintain data completeness. Ongoing updates are strengthening native support for high-novelty samples, making the platform increasingly robust for difficult plant genomes.
Our Workflow
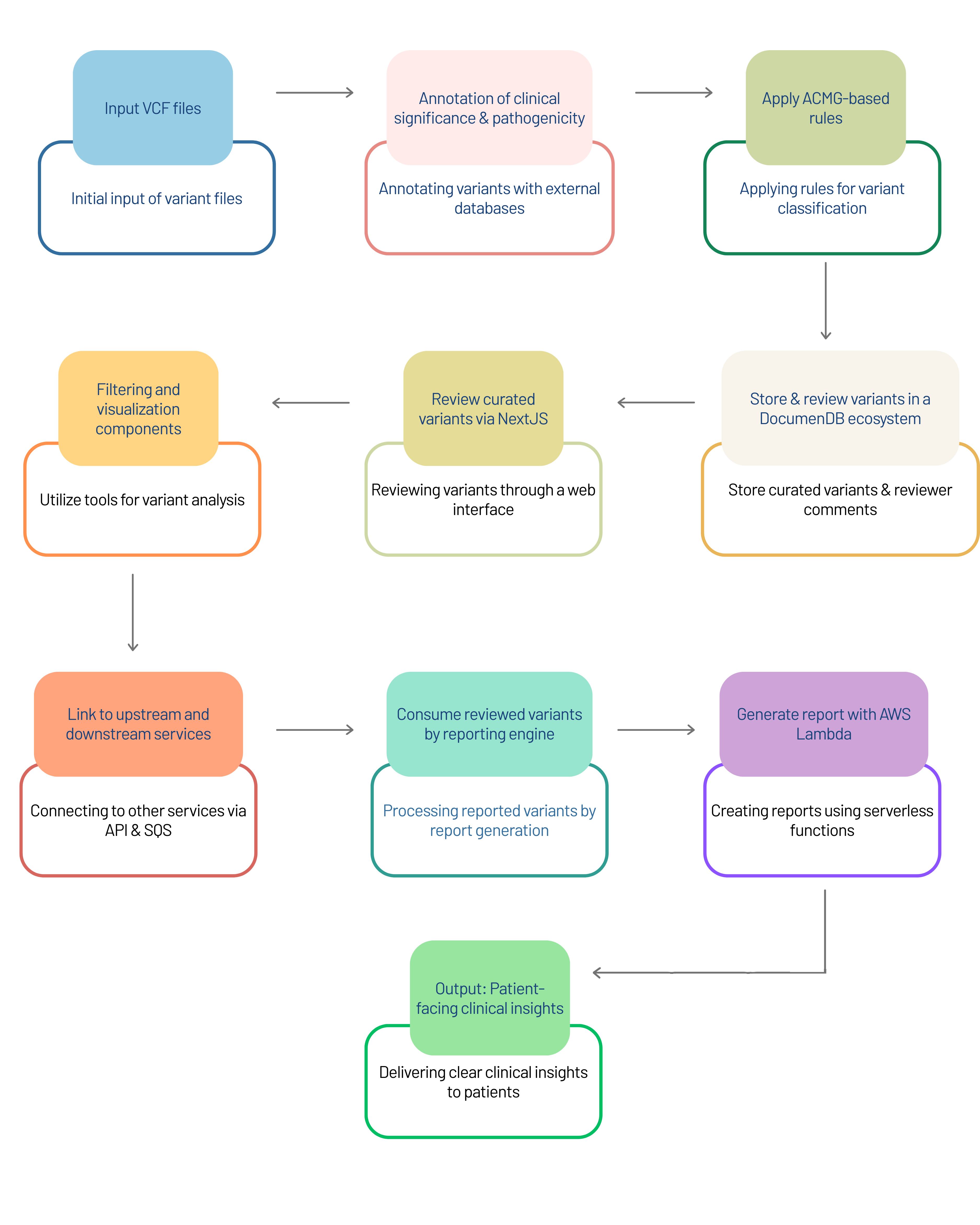
We have built supporting infrastructure that allows these analyses to run reliably at scale. The system is designed to handle complex datasets, automate key steps, and keep variant curation clear and traceable, ensuring results can be used confidently for downstream decision-making. This end-to-end automation, including dynamic regeneration of reports and a robust audit trail, reflects Strand’s ability to build scalable, reproducible, production-grade genomics pipelines that support both plant and clinical workflows (for example, a clinical workflow shared below).
If you are working in plant genomics and would like guidance or support, we would be happy to connect. Feel free to reach out at [email protected]
To explore more, watch our webinar on “How Millets Talk to Fungi: A Transcriptomic Perspective" and read our blog on “Identification of SNP Markers in the Pm4b Gene of Wheat”.
References:
- Kyriakidou M, Anglin NL, Ellis D, Tai HH, Bombarely A. Current strategies of polyploid plant genome sequence assembly. Frontiers in Plant Science. 2018.
- Chen F, Song Y, Li X, Chen J, Mo L, Zhang X, Lin Z, Zhang L. Genome sequences of horticultural plants: past, present, and future. Horticulture Research. 2019.
- Della Coletta R, Qiu Y, Ou S, Hufford MB, Hirsch CN. How the pan-genome is changing crop genomics and improvement. Molecular Plant. 2021.
- Pucker B, Irisarri I, de Vries J, Xu B. Plant genome sequence assembly in the era of long reads: progress, challenges and future directions. Quantitative Plant Biology. 2023.
- You FM. Plant Genomics – Advancing Our Understanding of Plants. International Journal of Molecular Sciences. 2023.
- Jayakodi M, Shim H, Mascher M. What Are We Learning from Plant Pangenomes? Annual Review of Plant Biology. 2024.
- Younas MU, Zuo S, Qasim M, Ahmad I, Feng Z, Korai SK, Zulfiqar U, Tukhtaboeva F, Ismoilov I, Malik T. Multi-Omics Approaches in Plant Biology: Decoding Agronomic Traits for Sustainable Agriculture. Plant Stress (Elsevier / ScienceDirect). 2025.