What's in a term? That which we call a nerve cell by any other name (neuron, perhaps?) would fire just as swiftly.
Amidst a web of tangles, where unharmonized data lies, bioinformaticians may beg to differ, Shakespeare.
In an increasingly data-driven world, biomedical scientists are inundated with unorganized data. In our last blog, we talked about data harmonization and how it helps integrate diverse datasets for enhanced consistency and compatibility. One of the main ways to begin harmonizing data is to meticulously tidy up the metadata.
This is where ontologies come to the rescue. So, what are they?
Ontologies are structured representations of knowledge that define concepts, relationships, and rules within a specific domain or across multiple domains. While taxonomies classify information into a hierarchical structure based on shared characteristics, ontologies hold both the structure and relationships between concepts. In very simple terms, taxonomies classify, and ontologies specify.
In biology, ontology is a formal representation of the hierarchical structure and relationships between biological entities, such as species, genes, proteins, and their attributes. Biological ontologies, or bio-ontologies, are used to organize and categorize biological knowledge systematically, making it easier to study, share, and integrate biological information. Bio-ontologies are also vital in supporting various tasks including data annotation, integration, analysis, and discovery.
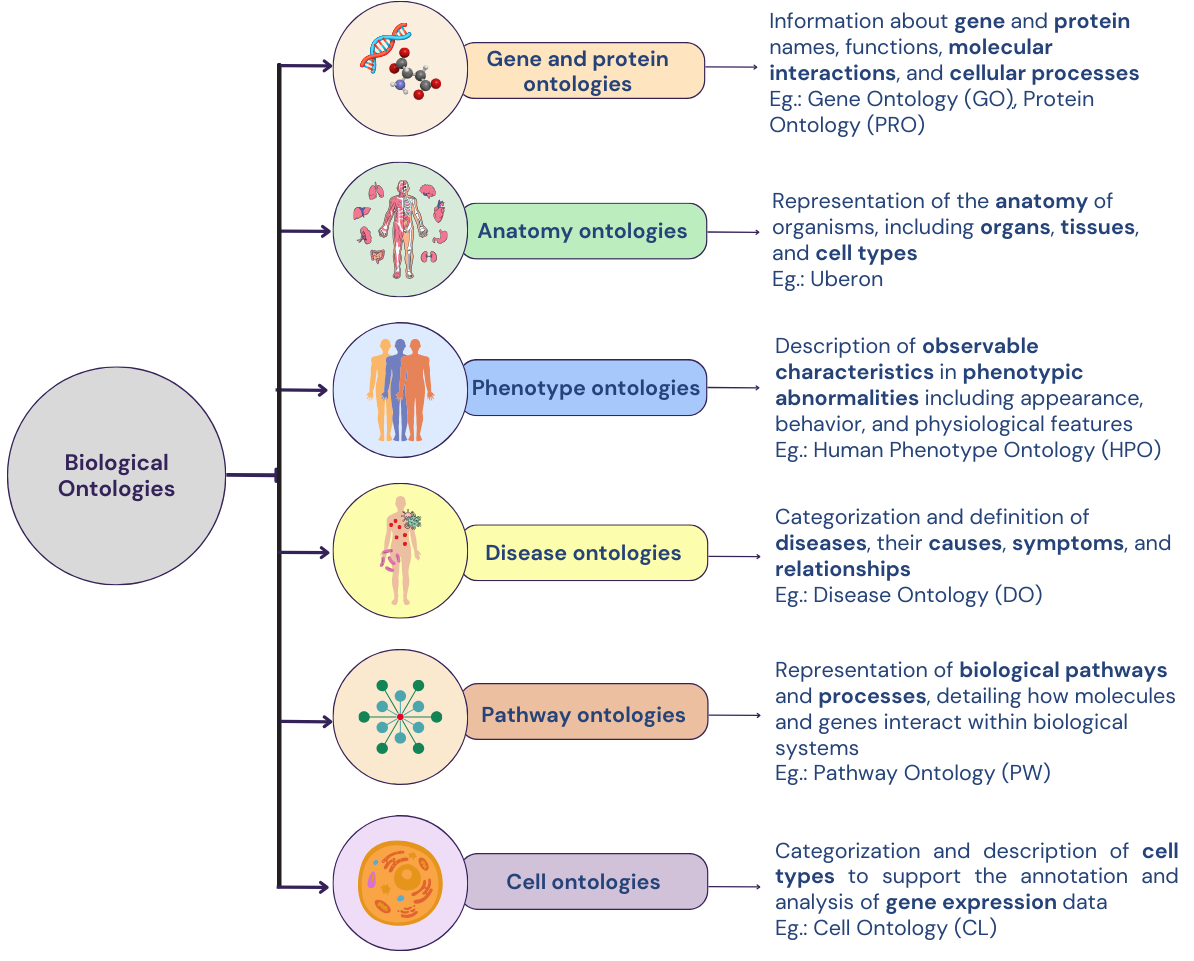
What do bio-ontologies look like?
Bio-ontologies are generally defined by either simple tree structures or directed acyclic graphs (DAGs).
In a tree structure, the hierarchy begins with a single root node, representing the broadest concept in the domain (i.e., the parent), and branches out into more specific concepts (i.e., the child) as you move down the tree.
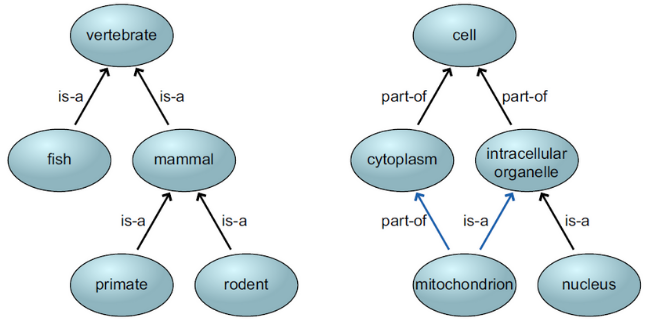
On the other hand, DAGs allow terms to be related to multiple broader terms, meaning that both parent and child nodes can have multiple connections.
Why are they important?
Bio-ontologies play a central role in the structured representation of biological knowledge. Their application spans genomics, proteomics, evolutionary biology, and bioinformatics, among other fields. Here are some important ways in which they contribute:
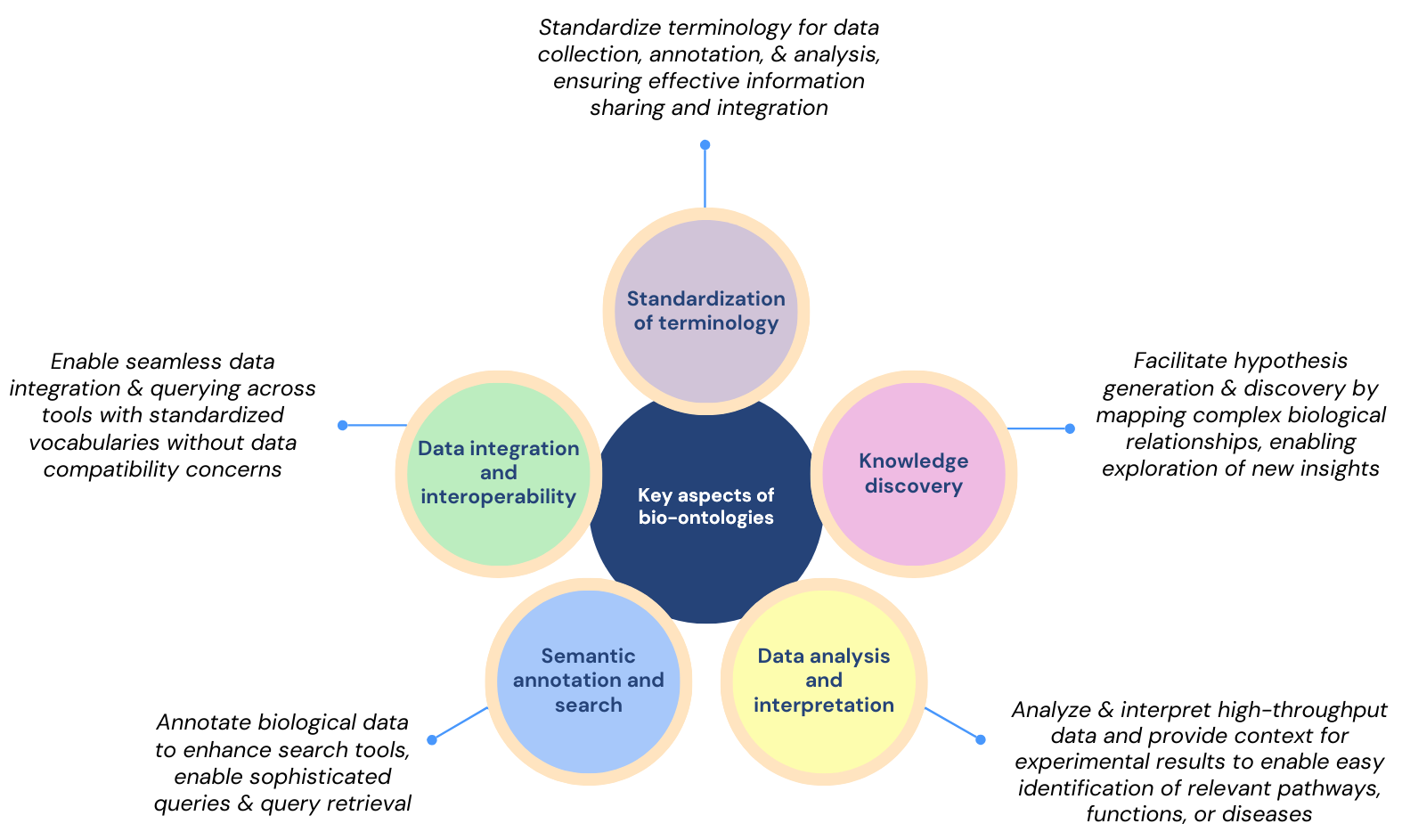
As described in the figure above, bio-ontologies standardize terminology, ensuring consistency and interoperability across research, and facilitate efficient data integration and analysis. By providing a framework for semantic annotations and standardized vocabularies, they enhance search capabilities, support high-throughput data interpretation, and promote knowledge discovery by mapping complex biological relationships, ultimately enabling researchers to generate new hypotheses and insights.
Thus, ontologies are indispensable in the context of biology for organizing and representing complex knowledge structures, as well as for ensuring semantic interoperability across diverse data sources and systems. Moreover, not utilizing bio-ontologies can give rise to several challenges, which we will address in Part 3 of the blog series.
For a more in-depth exploration of how we helped a big pharmaceutical company with data harmonization, read this case study!
You can find the rest of the blog series below:
- Part 1: From Chaos to Clarity: Harmonizing Data
- Part 3: Resolving Ontology Inconsistencies: Insights from Strand's Approach
- Part 4: Harnessing the Power of Harmonized Data: Strand’s Approach
References:
- Biological and Medical Ontologies: Introduction, Marco Masseroli, in Encyclopedia of Bioinformatics and Computational Biology, 2019
- Jensen LJ, Bork P. Ontologies in quantitative biology: a basis for comparison, integration, and discovery. PLoS Biol. 2010 May 25;8(5):e1000374. doi: 10.1371/journal.pbio.1000374. PMID: 20520843; PMCID: PMC2876043.
- Bard, J., Rhee, S. Ontologies in biology: design, applications and future challenges. Nat Rev Genet 5, 213–222 (2004).