Early disease detection is crucial to ensure early intervention and treatment, especially in cancer, where early diagnosis can greatly improve outcomes. However, traditional methods of disease diagnosis are slow, invasive, expensive, and often done at later stages. Liquid biopsies present a promising alternative, by providing a faster, non-invasive approach to detect and monitor cancer. By analyzing circulating free DNA (cfDNA) from a simple blood draw, they enable the identification of genetic changes, tracking of disease progression, and personalization of treatment, making cancer diagnosis more accurate and early intervention more feasible
One of the key aspects of cfDNA analysis is to study DNA methylation changes that are associated with cancers (as aberrant DNA methylation is observed in 70-90% of cancers). And, analyzing such epigenetic modifications (methylation profiling) and cfDNA fragment patterns (fragmentomics) can help identify the genetic alterations linked to disease presence and progression. However, the large volume of data generated makes CPU-based processing both time-consuming and costly.
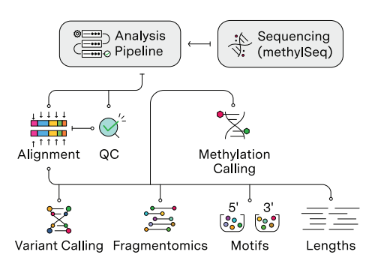
This is where cloud-native and GPU-powered solutions can make a difference. At Strand Life Sciences, we’ve tackled these very challenges with our cloud-native, GPU-powered genomics pipeline for cfDNA analysis that integrates methylation profiling, fragmentomics, and variant calling into one streamlined workflow.
Ensuring data integrity & accuracy at every step of the process, it can process up to 100 targeted methylation sequencing samples at 50 Gb per sample (100x coverage) in under 12 hours, with GPU-accelerated alignment bringing runtime down to below 10 hours—at a reduced cost.
The workflow includes extensive quality control checks of sequencing quality and alignment accuracy, including target enrichment metrics. Leveraging AWS for automated processing, it also offers scalability, reliability, and high-performance computing—ideal for large-scale genomic analyses.
Our pipeline consists of 4 different modules:
- Fragmentomics: Examines DNA fragment sizes, distributions, and patterns.
- Methylation Profiling: Tracks DNA methylation to study gene regulation and identify epigenetic markers linked to diseases like cancer.
- Variant Calling: Identifies genetic changes, such as mutations, insertions, and deletions.
- Quality Control (QC) Module: Monitors sample quality from start to finish, ensuring high-quality, reliable results for research and diagnostics.
As previously mentioned, traditional CPU-based tools struggle with scalability and speed, especially when handling large genomic datasets. Strand’s BWAMeth, optimized for GPUs instead of CPUs, on the other hand, delivers a 3.5x speed improvement over traditional CPU-based methods and reduces sample turn around time, accelerating cfDNA analysis. Not only does it match the accuracy of traditional CPU implementations, but it also reduces costs by 2.6x, offering a cost-effective solution for genomic analysis.

And hence, by simplifying workflows and enabling early disease detection through deeper insights from methylation and fragmentomics, Strand’s genomics analysis pipeline advances precision medicine and genomics research — with applications across diagnostics, cancer research, population genomics, and environmental & exposure studies.
Download a PDF version of this poster from our website or connect with us to explore how we can help you accelerate genomic data analysis!