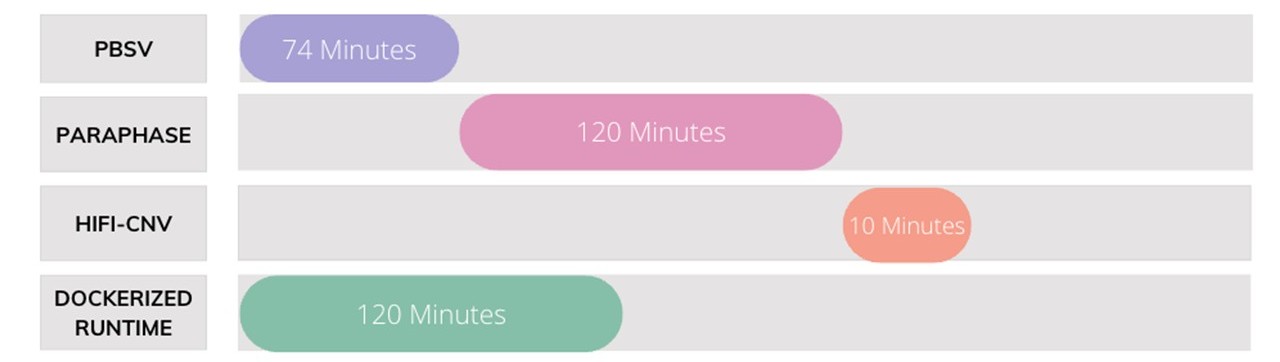
Run time can become a major bottleneck in analysis when working with large file sizes. So, when approached by a client to build a long-read analysis pipeline, we created a dockerized solution that cut down run time by 41%. Our pipeline produced a vcf containing over 50,000 variants from a 42 GB file in just 120 minutes. When run through the various tools individually, this same analysis took 204 minutes.
The client was working with unaligned long-reads obtained from a Pacific Biosciences machine, however, the analysis proved tedious and time-consuming. Each tool required separate setup, configuration, and resource allocation, leading to inconsistent environments, inefficiency in execution, and fragmented outputs.
We created a unified Docker-based solution that packaged these scattered workflows into one whole and optimized them such that it would be -$1.24 cheaper to run a typical 100GB WGS using this solution, and when run for one month, you would be able to run 62 more WGS samples as compared to running them through each tool separately.
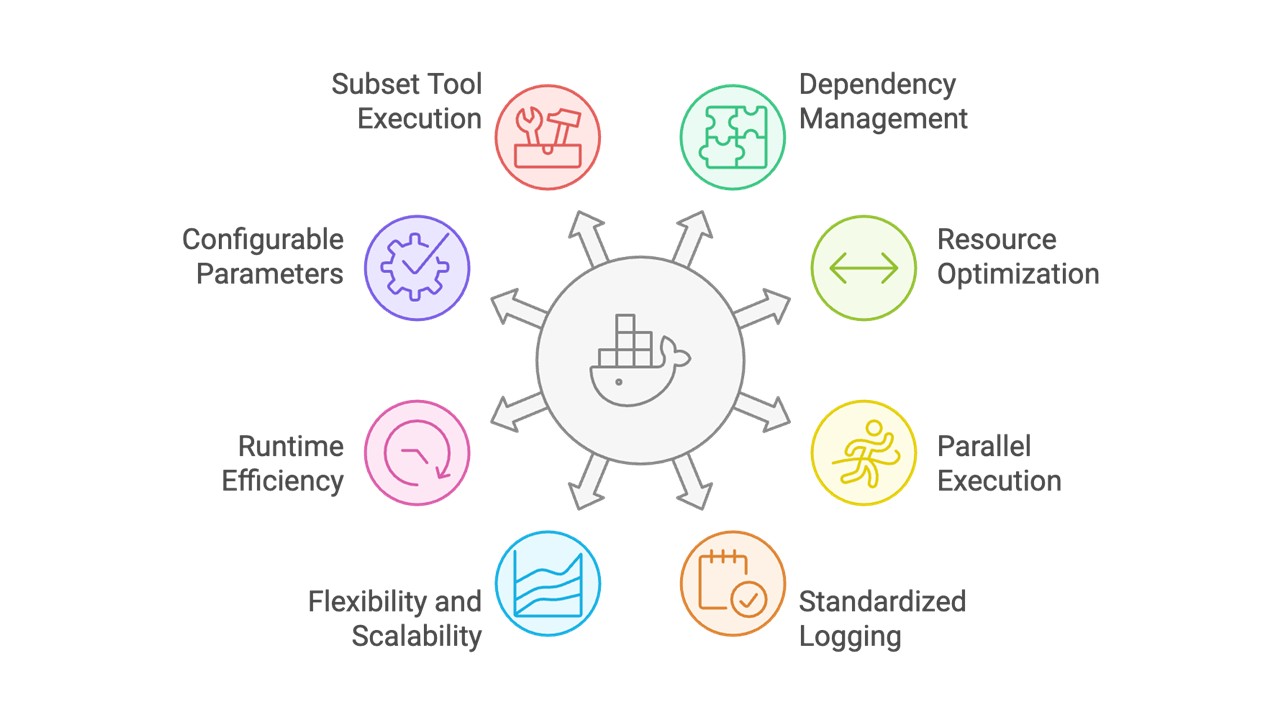
Through this Docker solution, we were able to:
- Manage different dependencies for each tool within a single Docker image without version conflicts
- Ensure tools can run in parallel without interference while handling interdependencies when they exist
- Implement standardized logging and robust error handling
- Ensure the Docker image is flexible for additional tools and scalable for larger datasets in the future
- Minimize Docker overhead and improve runtime efficiency to prevent the image from slowing down the analysis pipeline.
- Store parameters in config files for easy sharing and reproducibility
- Provide the option to run a subset of tools as per user’s need.
Our Docker solution streamlines tool execution, enabling consistent environments, parallel processing, simplified commands, and consolidated outputs* for efficient genomic analysis.
Contact us to learn more about how we can deliver a custom Docker solution for you!