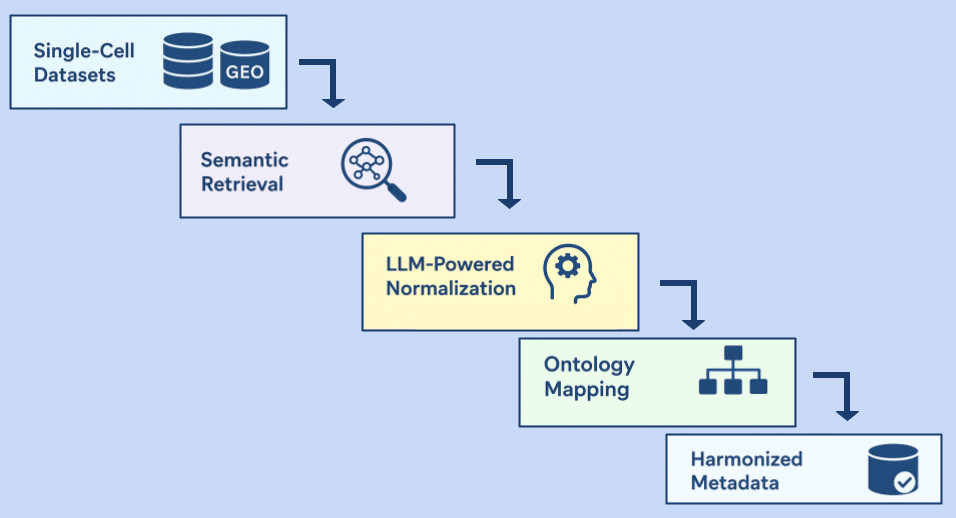
We are writing to introduce a novel computational pipeline designed to address a critical bottleneck in single-cell research: the automated normalization and harmonization of biomedical entities across heterogeneous datasets.
Our Retrieval-Augmented Generation (RAG) framework provides a robust solution for integrating disparate single-cell datasets from public repositories like GEO. At its core, the pipeline maps complex metadata fields—including diseases, tissues, and cell types—to their corresponding standardized ontologies, such as DOID, UBERON, and the Cell Ontology.
The key innovation lies in its architecture:
- Semantic Retrieval: We leverage state-of-the-art BioLORD-2023 embeddings, which excel at representing complex biomedical concepts, to perform highly accurate semantic searches for entity normalization.
- LLM-Powered Normalization: This retrieval mechanism is coupled with the advanced capabilities of GPT-4.1 and o3 reasoning model variants, which analyze the context and resolve entities with high precision.
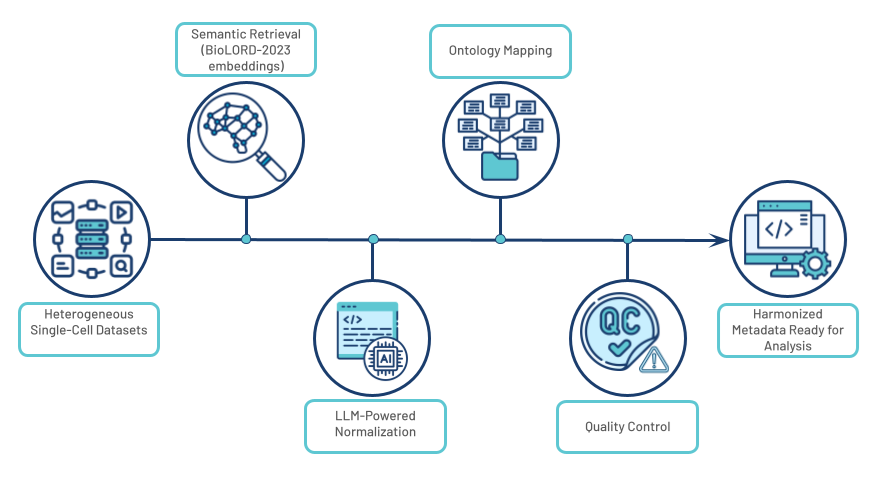
Fig 1. An overview of the RAG+LLM workflow pipeline
This RAG-based strategy has demonstrated superior performance over traditional methods, which often fail to capture the semantic complexity and contextual variability inherent in biomedical nomenclature. Our pipeline consistently achieves an average accuracy exceeding 95% across 15 key metadata fields, resulting in a three-fold reduction in turnaround time compared to manual curation.
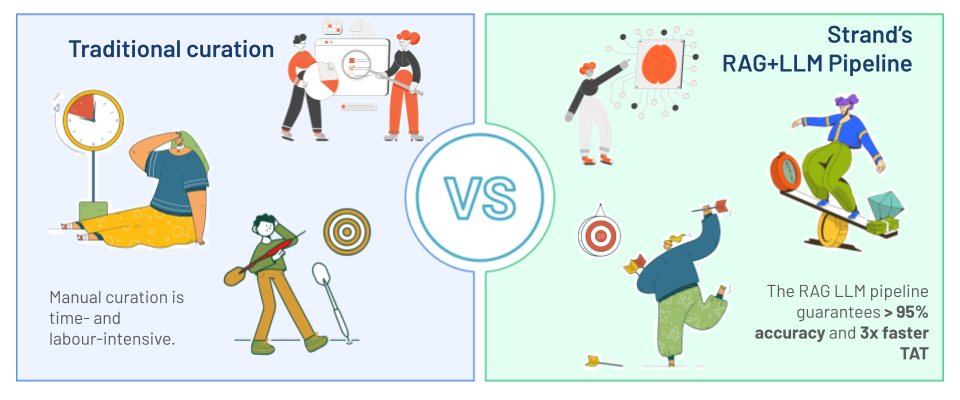
Fig 2. Strand’s RAG+LLM data harmonization and normalization pipeline offers substantial advantages over traditional curation methods
To ensure reliability, a comprehensive quality control system provides confidence scores for all automated predictions and flags ambiguous cases for optional manual review, guaranteeing both high-throughput scalability and scientific accuracy.
This approach enables researchers to harmonize large-scale single-cell datasets with unprecedented speed and precision, significantly accelerating comparative analyses and new discoveries in cellular biology.
If harmonizing complex biomedical data is a priority for your team, we would be delighted to share more detailed performance metrics and explore a potential collaboration.