Single-cell data is fast becoming a cornerstone of transcriptomic data analysis. Understanding not only what the gene markers are, but also which cell types they are being expressed in, can help improve our understanding of biological mechanisms at a more granular level. Large-scale single-cell transcriptomic datasets can provide deeper insights into the complex interactions and co-expression patterns among genes across various tissues. As this technology becomes more commonplace, the amount of data continues to increase, thus necessitating more automated analytical tools that can ingest and analyse this data at larger scales.
This is where NLP-derived foundation models such as BERT and GPT have begun to make an impact, especially with the advent of more specialised single-cell tools like scBERT and scGPT. In recent blog posts, we've been discussing the use of such foundation models in data harmonisation and curation, as well as for use in cell segmentation and spatial transcriptomics analysis.
We recently presented a suite of Foundation Model-based tools we developed to aid in single-cell analysis, called CytoRx AI, at Bio-IT World 2025.
This platform leverages different foundation models (including scBERT, scGPT, Geneformer, AttentionPert, and CancerFoundation, to name a few) to provide tools that can:
- Annotate Cell-Types
- Predict Gene Function
- Predict the Effects of Two-Gene Perturbations
- Integrate Multimodal Data
- Predict Cancer Drug Response
Computational Resources and Run Times for Each Task
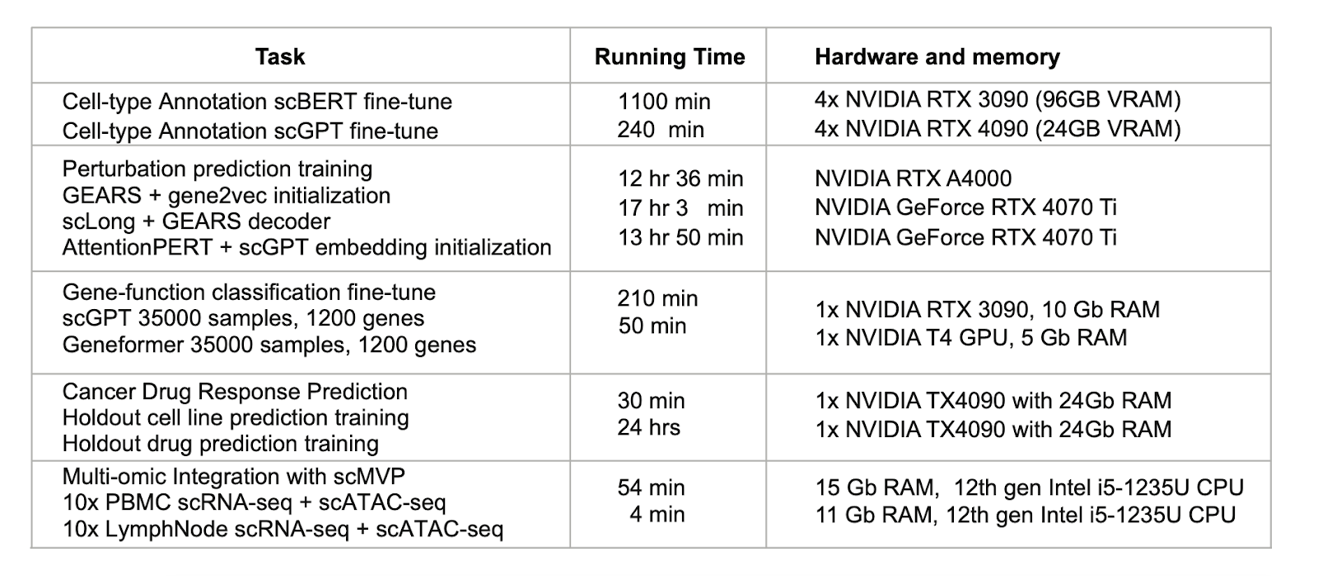
For each analysis, multiple foundation models are used, and their outputs are blended through stacked generalisation. For instance, cell-type annotation was done by fine-tuning scBERT and scGPT using a high-quality, curated Ulcerative Colitis dataset (Simillie et al., 2019) that was procured from our scRNA portal. The unified output from these models provided a more comprehensive picture of biological processes, disease and drug response, which, in turn, enabled the derivation of more detailed insights for target discovery via differential gene expression analysis. Foundation models also demonstrated improvements in a well-studied pancreas dataset (Chen et al., 2023).
When compared to conventional ML approaches, the optimized tools available on CytroRX AI demonstrated improved accuracy and outperformed previous methods for cell-type annotation (~7%, 33% improvements in macroF1), perturbation prediction (~10-20% decrease in mean square error), gene-function classification (~15% improvement in macroF1) and cancer drug response prediction (~15% improvement in Pearson Correlation) tasks.
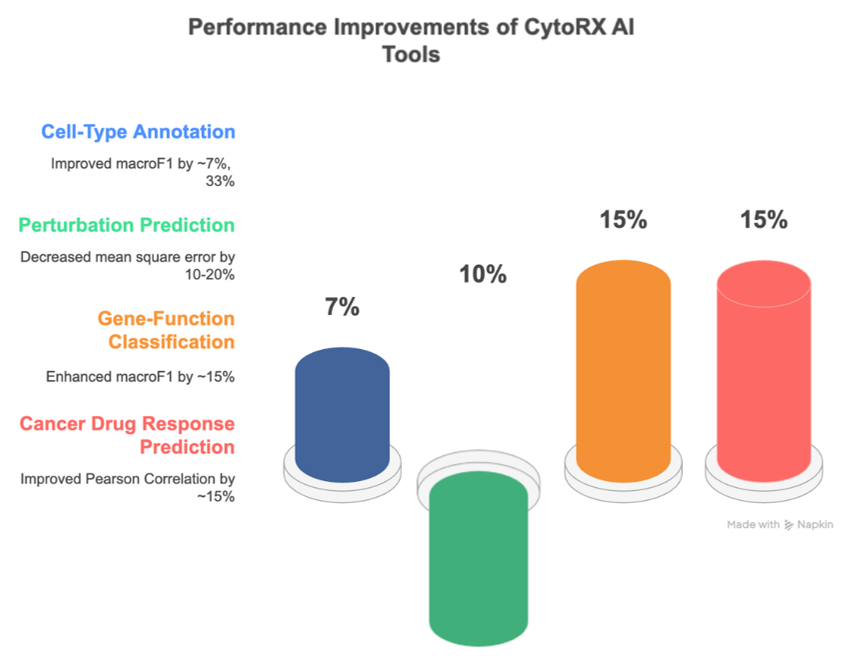
CytoRx AI illustrates how foundation models are not only feasible but also advantageous for single-cell analysis. By combining biological insight with state-of-the-art machine learning, it sets the stage for more accurate, scalable, and integrative tools in drug discovery. As the field advances, such platforms will be instrumental in turning single-cell data into actionable biological and clinical insights. To learn more about how you can access and use this platform, for additional information, or to download the poster, reach out to us at [email protected].