Nextflow is a workflow management software for developing scalable and reproducible workflows. In recent years, it has become increasingly popular in the bioinformatics space as it allows users to combine all of their various computational scripts into a single pipeline.
Some of the main advantages of using Nextflow are:
- Portability
- Reproducibility
- Parallelization
- Run time efficiency
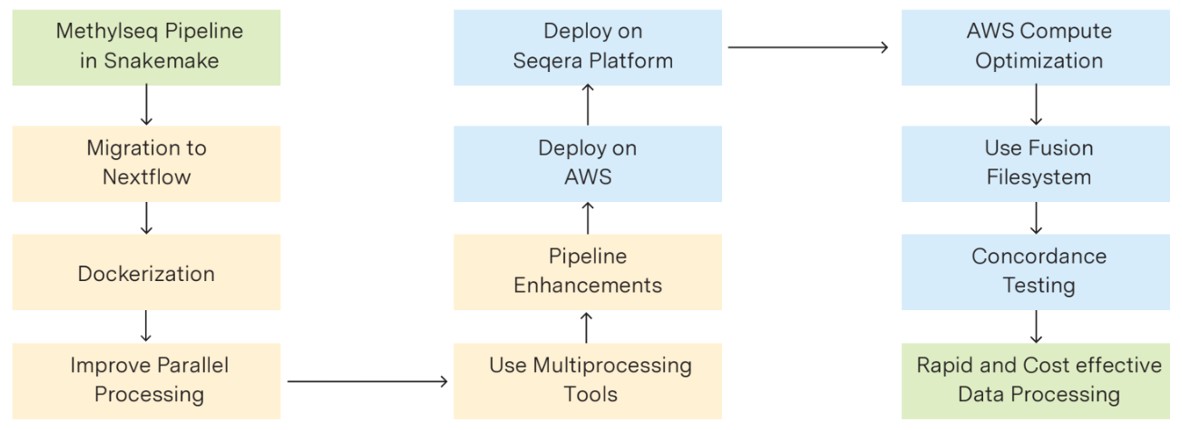
Here at Strand, we have leveraged these features to optimize our methylation pipelines, achieving over 80% reduction in turnaround time and more than a 90% increase in data processing volume for our customers. To summarize:
- The methylation pipelines were migrated to Nextflow and deployed on the Seqera platform, resulting in a >50% cost reduction and simultaneous processing of ~100 samples using AWS Batch and AWS ECS.
- The use of the Sentieon toolkit, which breaks large files into smaller chunks for simultaneous processing, helped achieve over 20% faster speeds compared to non-parallelized tools like Picard.
- The pipeline runtime was reduced from 8 days to just 1.5 days, enabling customers to increase processing from 1 batch per week (10 samples, 0.5 TB) to 20 batches per week (200 samples, 10 TB), ensuring the pipeline is no longer a bottleneck.
- The pipeline was dockerized to encapsulate dependencies, enhance scalability and reduce overhead.
Our bioinformatics team—Pavan Kotha, Neha Bhojani, Nishant Shekhar, Mayur Saini, Juhi Pandey, Ruthvik Bobba, and Jaya Singh—recently presented this work at Bio-IT World 2024. The PDF version of this poster is available here!

Overall, porting the pipeline to Nextflow, dockerizing workflow steps, deploying on the Seqera platform, and enabling multiprocessing led to significant improvements in scalability, efficiency, reproducibility, and cost-effectiveness.
Stay tuned for future updates on this project!