In the age of Big Data, it’s important to not only have good quality data but also well documented, easily accessible data. With a myriad of data sources both public and private, it is easy to run into issues when trying to access data from them. One way to tackle this is to use a harmonized solution with models and templates that can easily ingest this data as it is accessed, as discussed in a recent webinar.
To better understand the workflow that Strand has implemented to standardize the creation of such models, one of our senior content analysts, Sakshi Shinghal, spoke to Yasodha Kannan, a resident bioinformatician here at Strand.
Sakshi: Hi, I’m Sakshi Shinghal, a bioinformatician and geneticist who has worked with Strand for around 3 years. I recently obtained an MSc in Genomic Medicine from King’s College London, in conjunction with St George’s - University of London. I have experience in creating webpages for bioinformatics tools such as expression visualisation and variant analysis pipelines as well as developing variant analysis pipelines for different types of sequencing. I’m currently working at Strand as a senior content analyst, aiding in content development and marketing efforts.
Yasodha: Hi, I’m Yasodha Kannan S, a bioinformatics expert specializing in next-generation sequencing (NGS) data analysis and multiomics data harmonization. With a master’s degree in biomedical engineering from IIT Bombay, I bring a strong academic foundation combined with hands-on technical expertise to the field of bioinformatics.I have been part of Strand for about 6 years and my work encompasses a broad range of NGS data analysis, including DNA-seq, RNA-seq, and single-cell analysis, alongside developing customized bioinformatics pipelines for diverse research and clinical applications.
Sakshi: So let’s start with the basics, can you explain what metadata is and the role it plays in data harmonization?
Yasodha: Data is typically harmonized on the basis of its metadata. When trying to understand and access data, it’s the metadata that is used for these queries. Metadata itself is information about the experiment conducted. For example: tissue type, organism studied, age etc. Some other important fields are: which cell types were studied, disease condition etc. These help researchers narrow down their search for relevant datasets for their analysis.
Sakshi: I see, so metadata is really the data about our data. But, why is metadata curation and having standardized models important, especially when working with public datasets?
Yasodha: When it comes to public datasets, metadata heterogeneity tends to be an issue. This could be, for instance, inconsistencies in terminologies or missing metadata fields caused by diverse data sources and a lack of standardization. This can cause issues in data integration and AI/ML applications. Hence, harmonization of such metadata is required. Creating a standardized template or model to structure this data ensures consistency, compatibility, and accuracy across datasets.
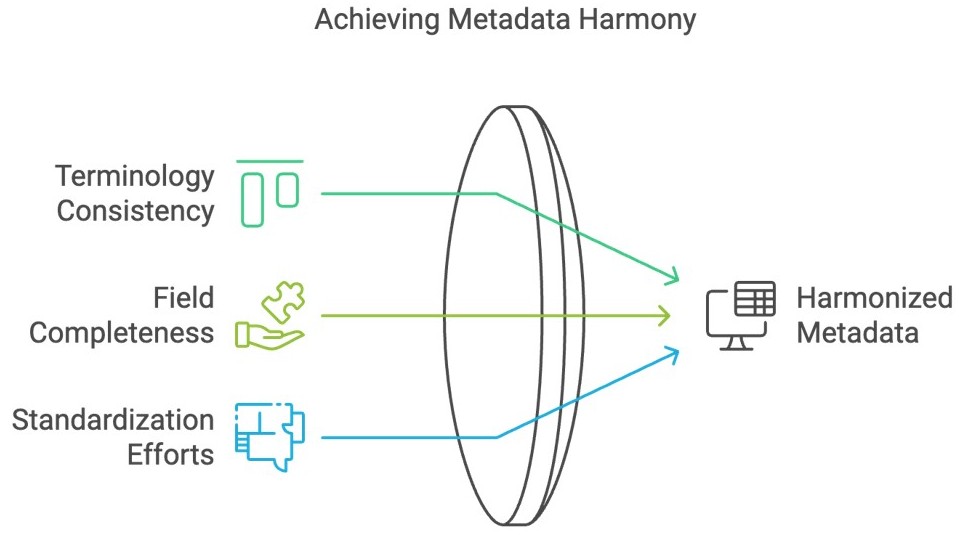
Sakshi: So data harmonization is really a crucial step, especially when working with large multi-modal datasets. Now, at Strand we have been harmonizing datasets for years, both for our own use and for other companies. Can you quickly go through the steps required to create a harmonized dataset?
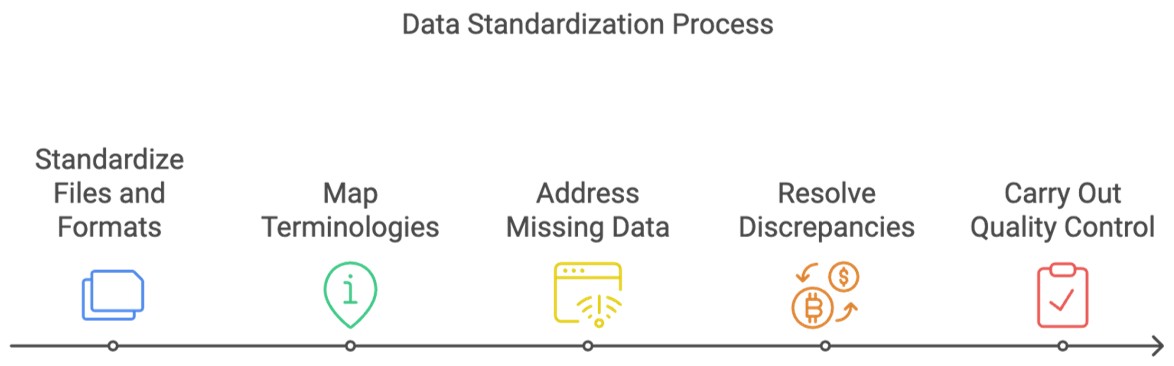
Yasodha: Sure, at a high level, first data files and formats have to be standardized. Then terminologies within these files have to be mapped. Following this, any missing data can be addressed and discrepancies can be resolved before we carry out final quality control checks.
Sakshi: Oh wow, that’s quite an interesting process, and rather involved. From my understanding we have created a standardized workflow for this harmonization. Could you talk me through it and how we would customize it for different use cases and data sets?
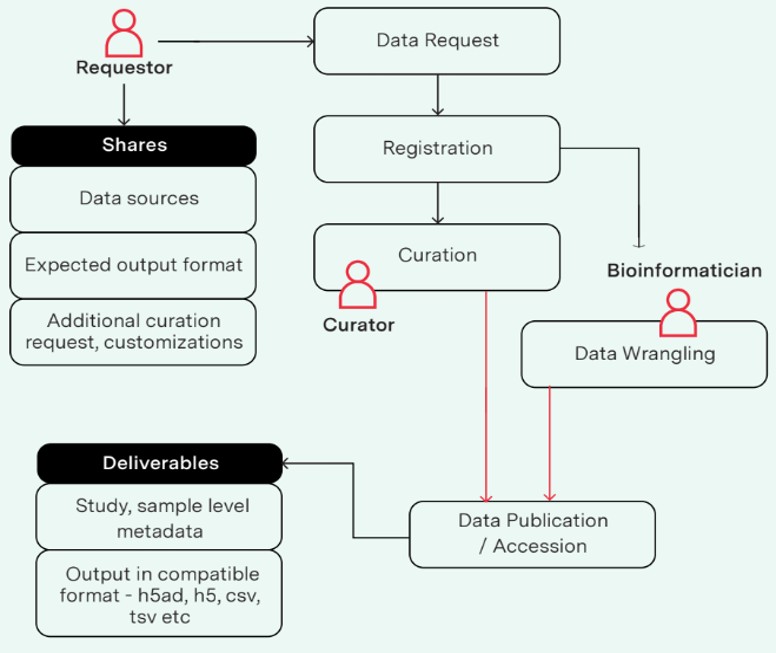
Yasodha: The process begins with a Requestor submitting a Data Request, providing key details such as:
- Data Sources: The specific datasets of interest.
- Expected Output Format: The desired format for the data (e.g., CSV, HDF5).
- Additional Curation Requests: Any specific customizations or additional steps needed for data preparation.
Once the request is registered, it progresses to the Processing stage. At this stage:
- A Curator reviews the available resources and completes the metadata template as requested.
- Simultaneously, a Bioinformatician conducts Data Wrangling, which involves cleaning, transforming, and preparing the data to meet the specifications outlined by the requestor.
After the curation and data wrangling are complete, the data is finalized and moved to the Data Publication/Accession stage. Here, the prepared dataset is made available, often through a repository or publication. The published dataset includes:
- Study and Sample-Level Metadata: Comprehensive information about the dataset and its context.
- Output in Compatible Format: The data prepared in the format requested by the requestor.
Sakshi: That’s fascinating! My understanding is we have recently introduced AI into this workflow? Could you tell me a little more about this?
Yasodha: To streamline ontology development and usage, we have looked into using LLMs to parse large text to extract potential terms, which are then mapped against existing ontologies using ontology-matching tools and AI models to identify matches or gaps. Unrecognized terms are flagged as candidates for review, validation, and potential integration, ensuring comprehensive and up-to-date ontology curation.
Sakshi: I see, so there’s a lot of space for AI in this process, would that mean we could move to a completely automated curation?
Yasodha: While AI plays a significant role in streamlining the curation process, moving to a completely automated system is not yet practical or desirable in most cases. Hybrid curation - a combination of automated tools and human expertise - offers the best of both worlds.
The process begins with automated extraction and validation, where AI-powered tools quickly parse and analyze large datasets, identifying potential terms, mapping them to existing ontologies, and flagging unrecognized terms. By leveraging AI, we can significantly reduce turnaround times (TAT), enabling faster delivery of curated datasets while managing larger volumes of data than would be feasible through manual methods alone.
However, certain aspects of curation, such as resolving ambiguities, addressing edge cases, and providing context-specific annotations, require the nuanced understanding and domain expertise of human curators. For instance, some terms may have multiple meanings depending on the scientific context, or there might be unique cases that deviate from the norm, which AI may not handle accurately.
By combining automation with expert review, hybrid curation ensures efficiency and scalability without compromising the accuracy and quality of the final output.
Sakshi: So that means while we use AI, it doesn’t negate the need for manual curation as well. Have we been able to try this hybrid model on any curation so far?
Yasodha: An excellent example of the hybrid approach in action is the curation of the 10x library prep field from free-text data for 3,000 datasets we did recently. Initially, manual curation was used to ensure high-quality extraction and structuring of this information. However, we enhanced this process by adopting a GPT-powered approach for ontology mapping of the same field.
The integration of GPT-4 proved highly effective, achieving an impressive 95% accuracy while demonstrating robustness in handling new and unseen data. By automating a significant portion of the workflow, we achieved a 2.5–3x improvement in ingestion turnaround time (TAT) compared to purely manual curation.
Sakshi: Ah, I see. That’s fascinating. Thanks so much Yasodha for taking the time to talk to me about this it’s been really great.
To learn more about our AI/ML efforts and some case studies visit us at us.strandls.com or reach out to us at [email protected]