Next-generation sequencing (NGS) has greatly advanced our ability to examine the genetic basis of diseases. In cancer research, NGS enables the study of disruptions in cellular signaling pathways that control growth, proliferation, and cell death, which are key factors in the development of malignant tumors. A deeper understanding of these pathways is crucial for developing targeted treatments that improve patient outcomes.
At Strand, our recent study utilized the Illumina TSO500 panel to analyze 2,015 in-house cancer samples to characterize their signaling pathways.
- This study classified SNVs, indels, CNVs, and fusions according to the Association for Molecular Pathology (AMP) tiering system.
- We focused on Tier 1 and 2 variants, mapping them to 11 literature-curated signaling pathways.
- By analyzing mutation profiles by cancer type and calculating co-mutation rates within pathways, we have been able to pinpoint critical alterations.
Our findings reveal that almost half of the samples showed alterations in more than one pathway, with an average of 1.87 pathways altered per case. Significant contributors to these alterations included KRAS in colon cancer, EGFR in lung cancer, and ERBB2 and FGFR1 in breast cancer. We identified the most frequently altered pathways as the receptor tyrosine kinase (RTK)/Ras, phosphatidylinositol-3 kinase (PI3K), homologous recombination repair (HRR), and the cell cycle (CC) pathways (Fig. 1).
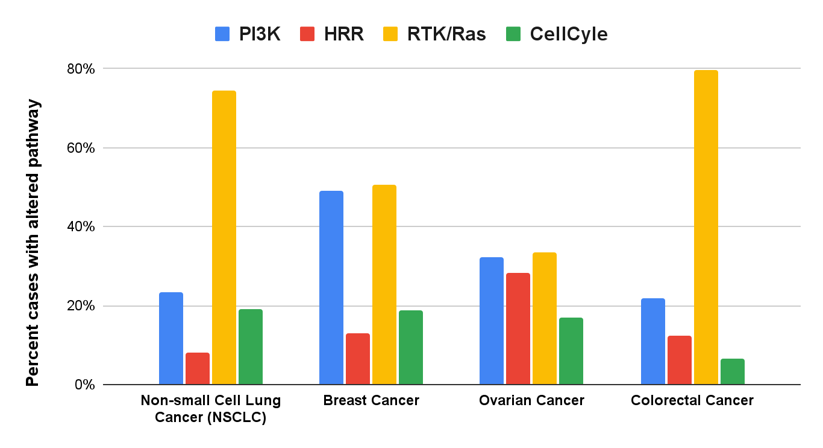
Fig. 1: Pathways activated across different cancer types.
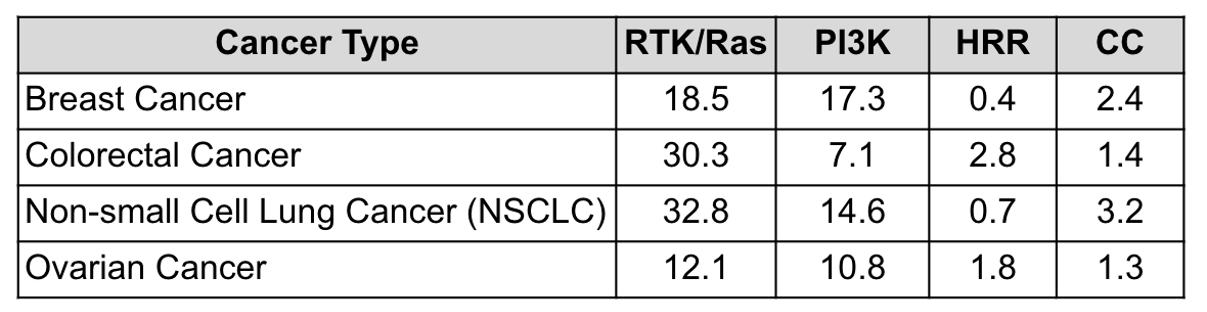
The study also revealed that multiple genes are altered within the same pathway. Co-mutation rates were determined by the percentage of cases showing alterations in two or more genes within a pathway (Fig. 2).
Why is this Study Important?
- Improved Understanding of Cancer Progression: By looking closely at the genetic data from 2015 cancer samples and mapping it to specific signaling pathways, we were able to better understand the step-by-step progression of cancer.
- Identification of Mutation Patterns: This extensive profiling not only identified 11 key pathways, but also highlighted the mutation patterns across 4 different cancers.
- Focus on Critical Pathways: With large-scale gene panels and NGS this research sheds light on tumor behavior and how responsive it might be to treatments, especially in lesser-known but vital pathways such as HRR and CC.
For more details regarding the study, check out our poster here.












