Cell-free DNA (cfDNA), particularly, cell-free tumor DNA (ct-DNA) has emerged as a novel diagnostic marker with untapped potential for improving clinical outcomes in cancer. Next-generation sequencing of ctDNA fragments, released into the bloodstream, offers a sensitive means of extracting valuable signals indicative of the tumor's underlying phenotype.
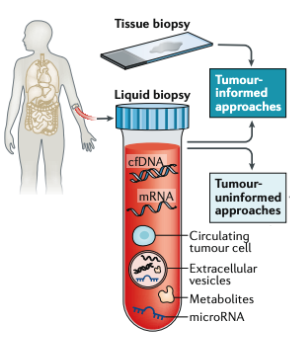
cfDNA Fragmentomics is an active area of research at Strand and our Bioinformatics team has developed an end-to-end pipeline, called FrAnaTK, for accurately analyzing cfDNA. Just about a month ago, we hosted a webinar delving further into cfDNA fragment signatures in liquid biopsies and showcasing our pipeline.
Eager to stay abreast of the latest insights, I recently attended a seminar at the Association of Molecular Pathology (AMP) conference in Utah, where Dr. Christina Lockwood from the University of Washington presented the most recent recommendations for working with ctDNA.
Building upon guidelines from 2015 and 2017, the latest version incorporates a meticulous review of literature from 2010 to 2020 (Lockwood et al., 2023). The survey was conducted on a total of 1228 publications. These studies included primary literature sources with a cancer-based focus on cfDNA in blood and excluded studies involving alternative fluids, and obsolete technologies (such as Sanger sequencing) as well as reviews and method-based papers.
The literature survey highlighted that thoracic and GI cancers were predominantly studied. Further, it sheds light on the critical role of pre-analytical variables such as blood volume, specimen collection tubes, storage conditions, and processing time on final analyte yield.
More importantly, it revealed that the lack of standardized reporting in certain areas, such as plasma separation and post-separation storage conditions, called for increased attention in future publications.
AMP’s recommendations, categorized into testing, reporting, and publishing, provide a clear roadmap for refining cfDNA assays. They emphasize the importance of addressing analytical requirements, understanding assay performance characteristics, and effectively conveying crucial information to both clinicians and patients.