We recently attended the AMP Conference in Salt Lake City, which brought together various streams of knowledge and talented scientists from all over the world.
One scientific talk I found particularly enriching was titled “Spatial Genomics in Solid Tumors,” presented by Dr. Andrew Gentles, an Assistant Professor in the Department of Pathology and Medicine at Stanford.
The talk brought to light the benefits of moving beyond our efforts to analyze tumor content and understanding the spatial organization.
Dr. Gentles walked us through the process of profiling the tumor microenvironment (TME) at scale, alluding to the current methods of flow cytometry and single-cell RNA sequencing, pointing out that each method comes with its own pros and cons.
The Gentles lab has developed a computational method for analyzing cell types present in bulk tissues, particularly estimating what cell fractions exist in a bulk mixture to understand the TME.
The lab’s efforts in this direction have yielded CIBERSORT (more recently referred to as CIBERSORTx), a widely used tool in the community for determining the cell type abundance and expression from bulk tissues (Newman et al., 2019). Given a bulk gene expression profile, the signature matrix, which is a reference dataset consisting of specific cell barcodes present in CIBERSORTx, is used to process the bulk mixture and estimate what proportion of the different cell types are present. Interestingly, CIBERSORTx is versatile and performs these deconvolutions at scale. Dr. Gentles's team has validated this method using 6500 samples of solid tumors derived from The Cancer Genome Atlas (TCGA) (Luca et al., 2021).
A fascinating aspect of CIBERSORTx is that it is not limited to cell type estimation but also helps identify specific transcriptional signatures of cells or cell states in cancers. These cell states are informative of prognosis and can thus potentially help in the management of cancer treatment.
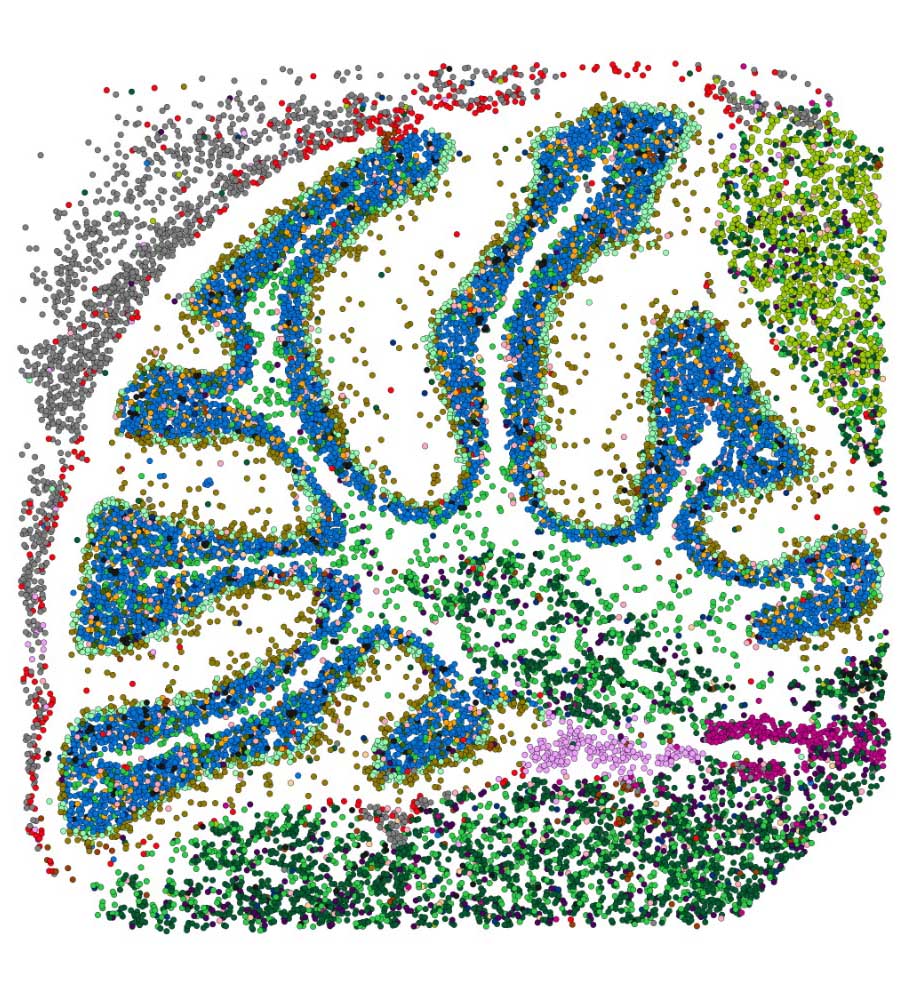
For those of us wondering how cell type deconvolution is related to the spatial organization of these cells in the TME, Dr. Gentles adds context by shedding light on the concept of Ecotypes. In essence, cell states don’t exist in isolation; they tend to co-occur in tumors. Just like individual cells, ecotypes are also prognostic of disease. Further, when their lab performed spatial transcriptomics analysis of solid tumor samples from TCGA, they discovered 10 ecotypes, each occupying spatially distinct niches.
Spatial biology is an exciting and fast-moving field, and at Strand, our teams are actively exploring methods and tools to leverage this technology for advancing cancer research. A significant milestone is our successful validation of the 10X Genomics Visium workflow in FFPE samples, culminating in our recognition as a Certified Service Provider for 10X Genomics. Additionally, our bioinformatics team has meticulously crafted an end-to-end spatial transcriptomics analysis pipeline, prioritizing speed and robustness, and in particular, cell type annotation at the single cell level.
For more insights into our spatial genomics endeavors, you can delve into the details here.