
Dockerized
PacBio Workflows
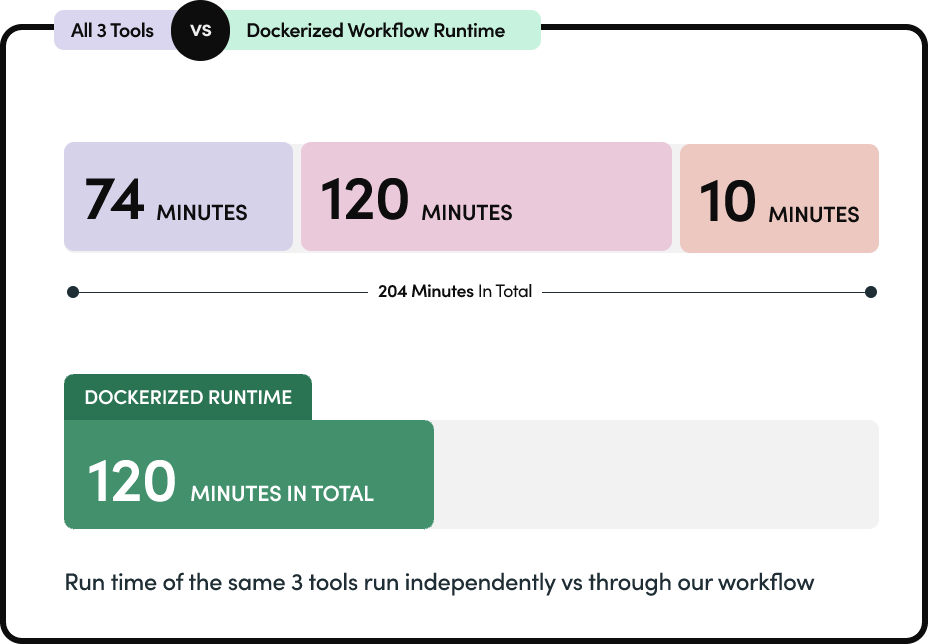
A unified Docker-based solution streamlines PacBio tool execution, enabling consistent environments, parallel processing, simplified commands, and consolidated outputs for efficient genomics analysis.


Managing different dependencies for each tool within a single Docker image without version conflicts.

Ensuring tools can run in parallel without interference, while handling interdependencies when they exist.

Option to run a subset of tools as per user’s need.

Implementing standardized logging and robust error handling.

Ensuring the Docker image is flexible for additional tools and scalable for larger datasets in the future.

Params stored in config files for easy sharing and reproducibility.

Minimizing Docker overhead and improving runtime efficiency to prevent the image from slowing down the analysis pipeline.

Minimizing Docker overhead and improving runtime efficiency to prevent the image from slowing down the analysis pipeline.
Advantages






